I just released a small R package that I have been working on for a while. The motivation for this package came from the observation that I kept on receiving questions about residual checks for GLMMs. The problem that people have is that they have fitted their GLMM, maybe they tested it for overdispersion and potentially corrected that, and now they plot their residuals and get something like this:
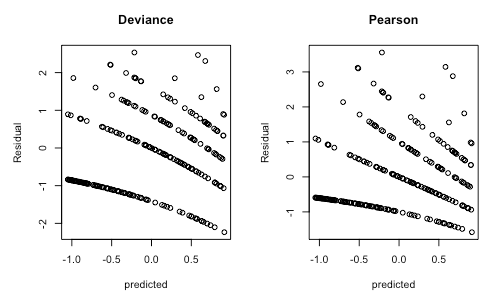
Deviance and Pearson residuals for a correctly specified Poisson GLMM
Is there a pattern that shows a dependence of residuals on the predicted value? Are the distributional assumptions of the fitted model correct? People are often concerned when they see plots such as the above because they learned in their elementary stats classes that residuals should look homogeneous. Well, that is the case for LMs, but not for GLMMs. When you work a bit more with GLMMs, you learn that inhomogeneous Pearson residuals are not necessarily a reason for concern. But even so, it is hard to tell if a GLMM is misspecified, just from looking at Pearson or deviance residuals.
The solution to this problem is well known, but a bit tricky to implement, which is presumably why it is seldom used. The goal of DHARMa is to change this and make residual checks for GLMMs as straightforward as for LMs.
The recipe is the following: for your fitted GLMM
- Simulated many new datasets with the model assumptions and the fitted parameters
- Define the residual for each observation as the proportion of the simulated observations that are lower than the observed value (see figure below)
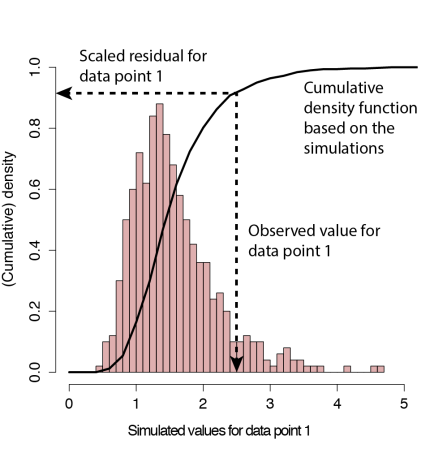
Definition of the scaled residual for data point 1, aka quantile residuals, aka the little sister of the Bayesian p-value. Simulations are performed with the fitted point estimate. The latter is the difference to Bayesian p-values, where the cumulative density of the posterior predictive distribution is used.
The resulting residuals (sometimes called quantile residuals) are scaled between 0 and 1. What is more important however, is that we have good theory about how they should be distributed. If the model is correctly specified, then the observed data can be thought of as a draw from the fitted model. Hence, for a correctly specified model, all values of the cumulative distribution will appear with equal probability. What that means is that we expect the distribution of the residuals to be flat, regardless of whether we have a Poisson, binomial or another model structure (see, e.g., Dunn & Smyth, 1996; Gelman & Hill. 2006).
Thus, you can simply plot your residuals, and if you see a pattern in the residuals, or the distribution is not uniform, then there is a problem in the model. To give an example: the plot below shows the DHARMa standard residual plots for a Poisson GLMM with underdispersion. The underdispersion problem shows up as a deviation from uniformity in the qq plot, and as an excess of residual values around 0.5 in the residual vs. predicted plots.
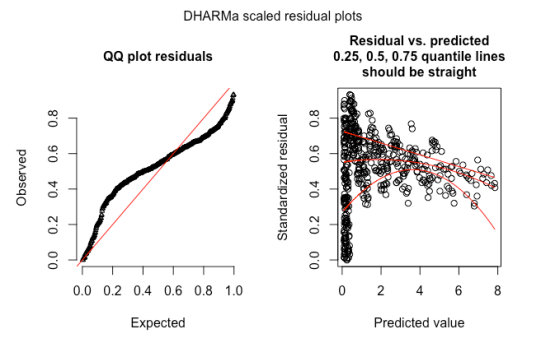
An example of a DHARMa residual plot for an underdispersed Poisson GLMM
That’s it basically. When we go into the details, there are a few further considerations. One is that the DHARMa package adds some noise to create continuous residual distributions for integer-valued responses such as Poisson or binomial. The other is that one can decide if new data is simulated conditional on the fitted random effects, or unconditional. The difference between the two is that for the first option, you look at whether your observed data is compatible with the model assumptions, conditional on the random effects being at their estimated values, while the second options re-simulates random effects and therefore implicitly tests their assumptions as well. I recommend to give it some thought which of your random effects you want to keep constant in the simulations. Some more hints are in the vignette.
The package also includes some dedicated test functions for
- Over/underdispersion
- Zero-inflation
- Residual temporal autocorrelation
- Spatial temporal autocorelation
Hence, no excuse any more to not check either of those for your fitted GLMMs. What is missing still are some dedicated tests of the random-effect structure. I’m planning to add this in the future.
p.s.: DHARMa stands for “Diagnostics for HierArchical Regression Models” – which, strictly speaking, would make DHARM. But in German, Darm means intestines; plus, the meaning of DHARMa in Hinduism makes the current abbreviation so much more suitable for a package that tests whether your model is in harmony with your data:
From Wikipedia, 28/08/16: In Hinduism, dharma signifies behaviours that are considered to be in accord with rta, the order that makes life and universe possible, and includes duties, rights, laws, conduct, virtues and ‘‘right way of living’’.
References
- Hartig, F. (2016). DHARMa: residual diagnostics for hierarchical (multi-level/mixed) regression models. R package version 0.1. 0. CRAN / GitHub
- Dunn, K. P., and Smyth, G. K. (1996). Randomized quantile residuals. Journal of Computational and Graphical Statistics 5, 1-10.
- Gelman, A. & Hill, J. Data analysis using regression and multilevel/hierarchical models Cambridge University Press, 2006


This is a great contribution! I’m looking forward to trying it, and I imagine the actual simulation process should be pretty quick for generating new residuals.
A couple things I was wondering about:
I was curious why you went with uniform residuals, rather than passing the uniform resids through the normal quantile function to get normally distributed residuals as in Dunn and Smyth. I’m not sure which is easier to visually detect deviations from the actual distribution, so both approaches seem fine, but my inner statistician always leans towards the normal distribution. 🙂
I was also wondering why you went with simulating new data, rather than just using the quantile function for the error distribution of the glmm that a user chose. I would think that, as long as you’re simulating new data from the same fixed effects and random effects as fitted by the model, the simulated distribution should just be the error distribution conditional on the mean value anyway. Does the simulation take model uncertainty into account, by drawing new fixed effects from the var/covar matrix?
Thanks again for putting this together! This does seem really useful, and hopefully helps people start to adopt a more error-statistical, model-testing approach with glmms.
LikeLike
Hi Eric,
thanks for the questions, great points!
About the uniformity: my statistical home is built more in Bayesian territory, and there you transform to uniformity (Bayesian p-values), so uniform seems more natural to me. I also think it’s a better choice because our eye is good at detecting inhomogeneities, but not deviations from normality.
That being said, I did consider the transformation to normality, because statistical tests that a user might want to apply on the residuals may require normality. As an example, I was concerned about the testTemporalAutocorrelation() function in DHARMa, which employs the Durbin-Watson test. I therefore implemented the normal transformation, you can access it via $scaledResidualsNormal in the simulated residual object.
I realized though that the transformation is not without problems – the main issue is that in many situations, it is quite likely to have all simulations lower or higher than the observed value, which leads to 0 or 1 in the current residuals, mapping to -Inf / Inf with qnorm. This could probably be fudged away, e.g. by adding some kernel density to the ecdf estimation, but who knows what that would do to the test properties, so in the end I decided that it’s safer to perform all tests on the uniform residuals (and the DB test seemed to perform fine on the uniform residuals).
Why not using the analytical quantiles / cumulative densities? Reason 1 is integer-valued distributions: imagine you expect 30% zeros, and you observe a 0 – on which quantile are you? Adding some noise essentially solved this, which is the point of the Dunn and Smyth paper. Reason 2 are the random effects – if you would use the CDF of your outer distribution, you residuals would be conditional on the fitted random effects. In DHARMa, the default is to re-simulate the random effects as well, which I think provides a stronger test of the model assumptions.
LikeLike
Hi Florian,
Sorry for the delayed reply; I’ve been in a big meeting for the past couple days, and wasn’t able to reply, but I appreciate the response!
Glad to hear you implemented both uniform and normal residuals. Do you know of any psychology literature on what kind of distributions it’s easiest to see a deviation from, and what kind of deviations people can detect visually? I tried Googling around, but couldn’t find anything. My instinct is that it should be easier to see changes in variance and maybe non-linearity with uniform residuals, but easier to detect outliers with normal residuals, as in the uniform case, large outliers would get contracted toward the center.
For the case of re-simulating the random effects: I wonder about the effect of this. It seems like this would always increase the variance of the simulated values, and therefore reduce how extreme a given residual is relative to its simulated distribution. In not sure I see when that would be useful for diagnosing modelling issues, as it would seem to obscure most patterns that would occur in the residuals. Is there a case you were thinking of where this would make a pattern of errors clearer or easier to detect?
I do generally think we should treat random effects as a higher-level residual as well; we often don’t test to see if the random effects follow the assumed distribution, or if there are relationships between our linear group-level predictors and the remaining random effects. However, I think that’s often best done by plotting the fitted random effects separately.
LikeLike
Interesting idea with the behavioral studies – I don’t know of any. I find it very plausible that general violations of the distributional assumptions are easier to detect in the uniform option, but yes, outliers may be less prominent to the eye – one would have to do an experiment.
About the random effects – my thinking is this: imagine you include an individual-level RE in a model with a missing quadratic predictor. The RE will fudge away the model misfit. However, now the individual random effect estimates will systematically depend on the missing predictor. This problem is pervasive in more complicated hierarchical models, e.g. state-space models always look great if you only look at the top level. If you re-simulate, you will see this. Of course, there would be other options to check this, most importantly to look at the RE estimates, but with re-simulating you get everything in one go. I therefore think it’s the better default option, even if you loose a bit power.
LikeLike
Hhhmm, good point about the individual-level random effects. Those are definitely important to simulate, since the random effect is essentially there as part of the error model anyway. For random effects where you have multiple observations per group, though, I still think you might be better off just treating the random effect as a higher-order residual, and look for patterns in the random effects.
For the behavioural studies: I feel like I have seen something about this, but I can’t find it for the life of me. Given how frequently we use residuals to assess model fit, it feels like such an obvious study to do, to actually test how well people can detect problems with a model using different types of residuals. I may poll the Rstats people on twitter to see if anyone knows about any study like this.
LikeLike
About the RE: hmm … I think I will do a few simulations to see how much power you loose if you re-simulate the RE. I wonder if people have given this much thought in general, also for Bayesian p-values.
If you do find out about the uniform / normal question, please do let me know.
LikeLike
If anyone is interested in further comments on the question of normal vs. uniform residuals – I have posted a few additional remarks here https://github.com/florianhartig/DHARMa/issues/39
LikeLike
There was a real need for a function like this, and this is a very nice solution, which I’ve been recommending to people who come to me with weird looking residuals plots. I’ve been using a simple visual version of the same approach (sim.residplot from https://github.com/pcdjohnson/GLMMmisc), which involves comparing the residuals-v-fitted plot to a simulated version, repeating a few times, and deciding informally whether the two look sufficiently similar. It also corrects the residuals from Poisson-lognormal GLMMs, which look nasty with the default resid and fitted functions.
Have you seen this paper?
Click to access Paper%2041%202001%20Elston%20Tick%20aggregation%20Parasitology.pdf
There are some similarities, e.g. using noise to get around the discrete response problem (Fig 2). It also looks at the distribution of the REs — I guess there’s less of a need for simulation here as the assumed distribution is always normal (in lme4, at least).
I’ll look forward to further developments!
LikeLiked by 1 person
Hi Paul,
thanks for the encouraging words, and the reference as well, which I hadn’t known before. I guess there have to be even more applied papers that use this method, it’s a known concept after all, especially in Bayesian Analysis.
How to deal with the random effects is still an issue for me, see also the discussion above. The random effect model assumes, but importantly also ENFORCES normality of the REs, in the sense that even if the underlying distribution of the REs is not normal, the estimates may look normal after the fit. Or at least more normal than they really are. So, performing a normality test on the RE will not be optimal in terms of power, and simulation could solve that issue – whether that matters in practice is yet another question.
Besides this more theoretical point, however, there is also a practical issue. Re-simulating the REs tests all assumptions of the model in one go. If you don’t do that, you would not only have to test your REs for normality, but also for correlation with predictors, residuals, space and time, because all this can indicate that the REs mask an issue in your model. Imagine, for example, that you have a spatial effect that is not accounted for by the model. An individual-level RE added to account for overdispersion may absorb this spatial effect so that nothing appears in the normal residuals – you would only notice if you plot the RE estimates against space, but who does that? So, I’m still thinking, but I do lean towards keeping the current default of DHARMa to re-simulate the REs as a default.
LikeLike
Hi Florian,
I’ve been trying out your package, and it seems to be working quite well, except when I try to use it on glmms with a gamma distribution. The simulationOutput function gives the message
“Warning message:
In rgamma(nsim * length(ftd), shape = shape, rate = shape/ftd) :
NAs produced”
and plotSimulatedResiduals produces very bizarre looking plots.
I tried to simulate some data to understand why, but I can see that the createData function doesn’t support a gamma distribution. Is this a feature you could potentially add?
LikeLike
Hi Christine,
thanks for reporting this – I have just checked the DHARMa residuals for a Gamma with glmer and didn’t encounter any problems.
Could you provide me with a bit more details about what you are doing? Ideal would be to create an issue here https://github.com/florianhartig/DHARMa/issues with a self-contained R script that produces the error, so that I can run the code and see what the problem is.
Thanks,
Florian
LikeLike
For future reference, this was discussed and solved in https://github.com/florianhartig/DHARMa/issues/8.
LikeLike
Pingback: Bayesian model checking via posterior predictive simulations (Bayesian p-values) with the DHARMa package | theoretical ecology
This seems like a great package. I’m familiar with these models in general, but I’m new to simulation to handle residuals. However, one thing I’m curious about is how researchers might go back to their original data set and transform or exclude cases based on the results of the simulation. Perhaps as I dig more around it will become obvious, but a description of that would make your package even more useful. Thank you so much for creating this.
LikeLike
Hi Steve, how to deal with that is described in https://cran.r-project.org/web/packages/DHARMa/vignettes/DHARMa.html
LikeLike
Hi Florian!
I’m trying the DHARMa package for a GLMM. To note in advance, I’m not a professional in statistics.
So, I’m running into following problem:
I use the simulateResiduals to do the residual analysis of my GLMM. Then the following is returned:
“Unable to calculate quantile regression for quantile 0.25. Possibly to few (unique) data points / predictions. Will be ommited in plots and significance calculations.
Unable to calculate quantile regression for quantile 0.5. Possibly to few (unique) data points / predictions. Will be ommited in plots and significance calculations.
Unable to calculate quantile regression for quantile 0.75. Possibly to few (unique) data points / predictions. Will be ommited in plots and significance calculations.”
Is this a problem and if yes how to solve it?
Many thanks in advance,
Saskia
LikeLike
Hi Saskia, the error message seems clear to me – what exactly is your question / doubt? Also, if you have further questions about DHARMa, the best place is to create a issue on the DHARMa GitHub repo.
LikeLike
Hi Florian!
Thank you for your reply. Unfortunately, the error message is not clear to me.
I thought it is because of the limited observations the GLMM is based on (~140 oberservations) and the maybe too many variables (about 5 or even more) of the model. However, if reducing the variables in the model to only one fixed and one random effect, I still run into the same error message. So I probably don’t understand why I run into it.
LikeLike
Maybe I got it now. Sorry for the second posting. The issue of not having enough data points for doing quantile regression is not dependent on the number of variables within a model?
However, the points within the plot residuals vs. predicted should be as randomly distributed as possible, right? So this is similar to the build-in LM diagnostic plots in base r, particularly the first one with residuals vs fitted? This is how I understood it after reading about GLMM, DHARMa, etc.
LikeLike
Hi Saskia, sorry for the late reply – I don’t check this blog very frequently, for faster responses to questions regarding DHARMa, feel free to use the issue tracker at https://github.com/florianhartig/DHARMa/issues
Anyway, regarding your questions
a) whether the quantile regression works depends indeed mainly on the number of data points, but also on their distribution (and not on the predictors). I would have to see your residuals to see why the regression lines fail in this case. Maybe a large number of your values has the same x or y value, which would make it harder to fit a quantile line.
b) yes, the DHARMA residuals are similar to the LM, just that the expected distribution if the model is correct is uniform, not normal. See the package’s vignette for a more detailed discussion.
LikeLike
Hi Florian!
It could also be related to using a quite old version of R. I updated it and now it shows the regression lines for quantile regression.
Thanks a lot!
Best,
Saskia
LikeLike
This is so helpful, thank you! ❤
LikeLike