Our new review on statistical inference for stochastic simulation models is now available at Ecology Letters.
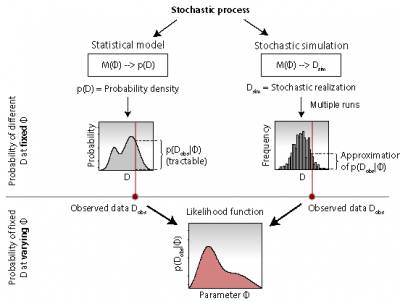
The paper discusses how approximate likelihood functions can be generated from stochastic simulations. Such simulation-based likelihood approximations are a very general and powerful tool because they allow us to apply statistical methods to a large class of models such as individual based models (IBMs), agent based models (ABMs) or metapopulation models for which likelihoods (i.e. the probability of obtaining the observed data from the model) could in principle be calculated, but this calculation is not tractable, i.e. can not be done analytically or numerically in an efficient way. Our main focus is on methods such as Approximate Bayesian Computation (ABC) or indirect inference, but we also discuss alternative approaches such as pattern-oriented modeling (POM) or generalized likelihood uncertainty estimation (GLUE).
Figure: Inference for statistical models vs. stochastic simulation models. From repeated realizations of a stochastic simulation model, it is possible to generate approximate likelihood functions. Based on such an approximation, a stochastic simulation model can be treated like any other statistical model.
Pingback: Responses of Late Quaternary megafauna to climate and humans « theoretical ecology
Pingback: struggling with problems already attacked | Hypergeometric